We research and develop light-based solutions for questions and challenges in the fields of health, environment, medicine and safety. Solutions that make life safer, healthier and cleaner.
Unfortunately, this video can only be downloaded if you agree to the use of third-party cookies & scripts.
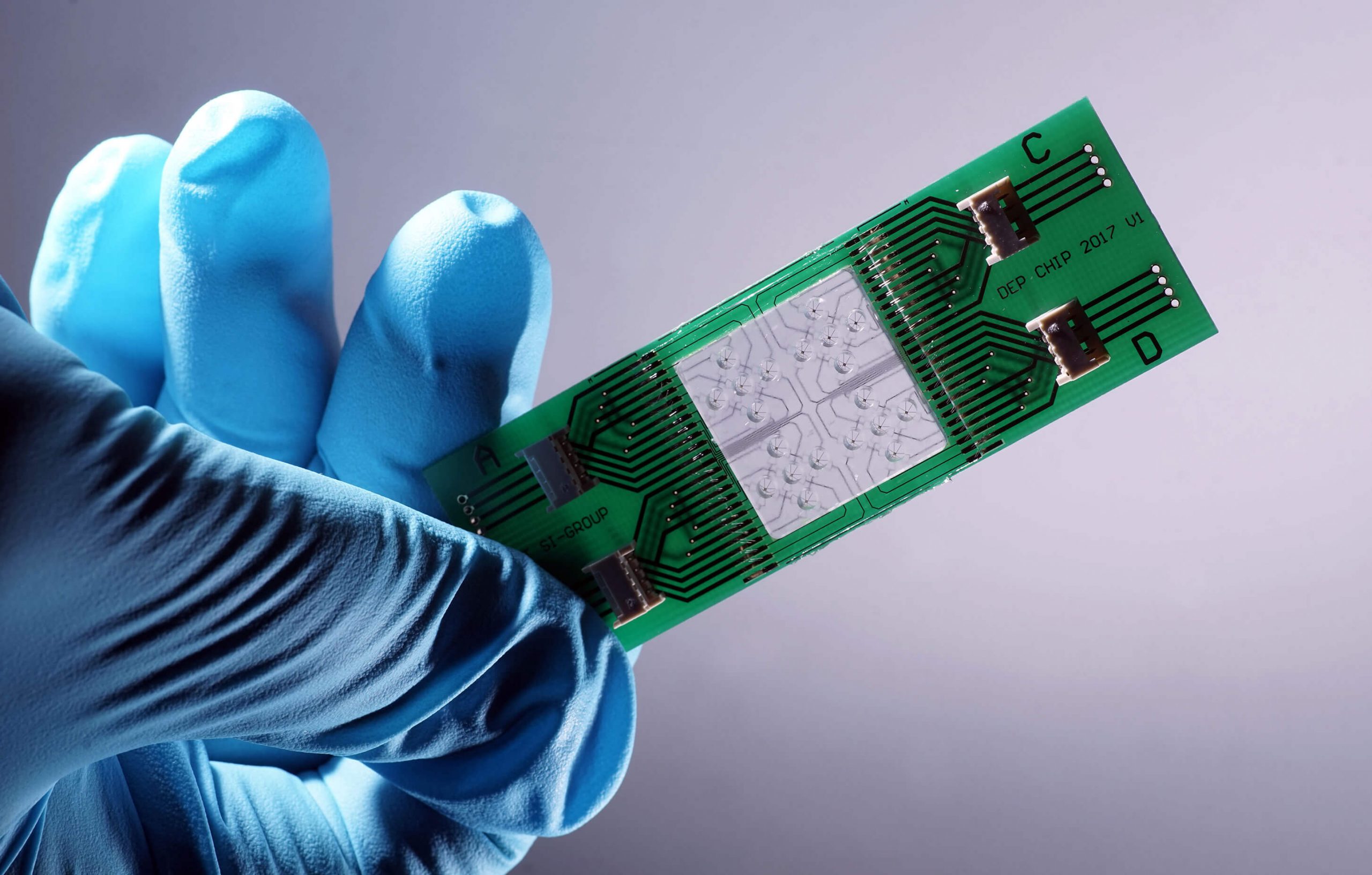
The focus of the scientific work at Leibniz IPHT is on optical health technologies and is oriented towards current medical challenges and needs. The research activities are divided into six thematic areas: Biomedical Microscopy and Imaging, Multiscale Spectroscopy, Ultrasensitive Detection, Special Fiber Optics, Nanoplasmonics, and Bioanalytical and -medical Chip Systems. They all have access to the key technologies available at the institute, such as micro/nanotechnology, fiber technology, systems technology and artificial intelligence methods.
Research Departments
Biophysical Imaging
Prof. Dr. Christian Eggeling
Clinical Spectroscopic Diagnostics
Prof. Dr. Ute Neugebauer
Fiber Photonics
Prof. Dr. Markus Schmidt
Fiber Research and Technology
Prof. Dr. Tomáš Čižmár
Functional Interfaces
Prof. Dr. Benjamin Dietzek-Ivanšić
Junior Research Groups
Leibniz IPHT is committed to making research results effective for society; for example, as a basis for technological innovations and political decision-making processes. The involvement of future users in the research process promotes the effective transfer of technologies, such as for medicine. Researchers at Leibniz IPHT conduct science from basic research to the development of a prototype — from Ideas to Instruments.