- Home
- Research
- Spectroscopy and Imaging
- Research results
- Surface-enhanced Raman Scattering of Cell Lysates for Improved Tumor Detection
Surface-enhanced Raman Scattering of Cell Lysates for Improved Tumor Detection
12.04.2018
Surface enhanced Raman scattering (SERS) is a powerful technique which can be used for cell identification and differentiation. A SERS approach with silver nanoparticles previously developed for bacterial cell detection was transferred to tumor cell lysates. The combination of SERS and microfluidics increased the throughput and accuracy of cell discrimination.
By: Christoph Krafft // Mohamed Hassoun // Jürgen Popp
According to worldwide cancer statistics from 2012 (www.cancerresearchuk.org), 14.1 million new cases and 8.2 million cancer-related deaths have been estimated per year. The number is expected to increase towards 23.6 million new cases of cancer each year by 2030. The importance of this topic is evidenced by the fact that, statistically, one in three Europeans will develop cancer within his lifetime. Raman spectroscopy has been shown to detect cancer tissues and cells without labeling or other preparation. However, the intensities of Raman signals depend on the weak process of inelastic light scattering. Surface enhanced Raman scattering (SERS) with noble metal nanoparticles can increase the intensity of Raman bands by several orders of magnitude. The enhanced Raman signals enable the detection of analytes at lower concentrations, the reduction of exposure time, and better throughput. The recognition of tumor cells by immune-SERS markers in a microfluidic flow at 25 ms of exposure time has recently been shown (1). However, the protocol for preparing immune SERS markers was very complex. Here, silver nanoparticles (Ag-NPs) that can easily be prepared in a single step process according to a protocol by Leopold and Lendl were applied to detect tumor cells.
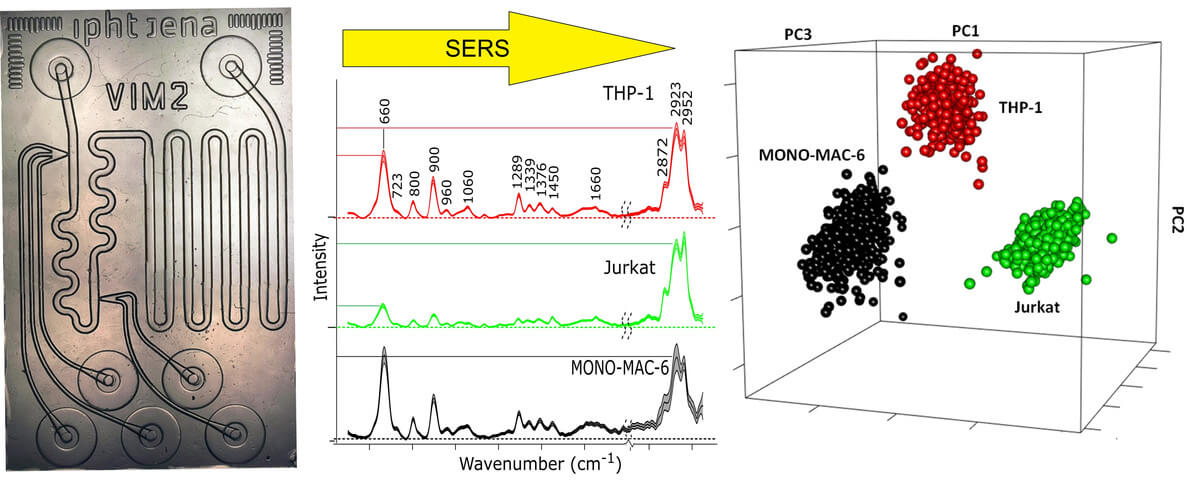
Our approach starts with the lysis of cells by an ultrasonicator probe. The homogeneous cell lysate contributes to high reproducibility because all intracellular biomolecules are able to interact with nanoparticles. As Ag-NPs show plasmon resonance in the range between 400 nm and 500 nm, potassium chloride (KCl) was added to induce aggregation and shift the plasmon resonance towards 800 nm for SERS with 785 nm of laser excitation. The approach was first demonstrated in ninety-six well plate cuvettes where the cells of two liver cancer cell lines, HepG2 and SK-Hep1, one pancreatic cancer cell line, Capan-1, and one breast cancer cell line, MCF7, were mixed with Ag-NP and KCl. The laser was focused on the surface of the mixture and SERS spectra were collected. The experiments were repeated using six batches for each cell line to demonstrate reproducibility. Pronounced spectral variations were found between cell lines, whereas the standard deviation between SERS spectra of different batches were small. A support vector machine (SVM) was trained to classify the four cancer cell lines with high accuracy above 96%. More details are reported in (2).
Then, this approach was transferred to a microfluidic lab on a chip (LOC), which was made from a quartz wafer for compatibility with 785 nm of laser excitation. LOC experiments make it possible to accurately generate droplets in a flow of mineral oil by mixing nanoliter volumes of Au-NPs, KCl, and cell lysate. Reproducible SERS spectra of thousands of droplets were collected at a defined position of the chip with one second of exposure time. Three batches of three leukemia cell lines, MONO-MAC-6, THP-1, and Jurkat, were prepared, lysed via ultrasonication and injected into the chip. SERS spectra with spectral contributions from the separation medium oil were removed from the data set before SVM classification. The data was divided into a training set composed of spectra recorded from two batches of three cell lines and a test set composed of spectra recorded from another batch of the three cell lines. The test set was then classified by the SVM model developed using the training set. Based on classification results, the accuracy, sensitivity, and specificity of the SVM model for identification of THP-1, Jurkat, and MONO-MAC-6 were calculated and compiled in a confusion matrix. All values were better than 99%, which is consistent with more accurate mixing, less standard deviation, and improved reproducibility in (3) compared to the previous paper (2).
Subsequent research work will include non-tumor cells as a control, promote cellular uptake of Ag-NPs for single cell analysis, and detect the fraction of cancer cells in mixed populations.
Funded by: DAAD
