- Home
- Research
- Nanoscopy
- Research results
- Hybrid Protein Nanofibers – An Analysis on the Nanoscale
Hybrid Protein Nanofibers – An Analysis on the Nanoscale
28.03.2019
During the self-aggregation of proteins, elongated twisted structures can form that exhibit high biocompatibility and interesting mechanical properties. If such nanofibers are generated from a protein mixture, an analysis on the nanometer scale using tip-enhanced Raman spectroscopy (TERS) can directly determine whether both proteins are contained in the fibers.
By Tanja Deckert-Gaudig // Volker Deckert
The self-aggregation of proteins into so-called amyloid fibrils is usually associated with neurodegenerative diseases. However, the twisted structures of amyloid fibrils are biocompatible and very strong. This can be used for the development of new drug transport systems and the production of biosensors.
Based on protein blends, hybrid nanofibers can be generated that combine the specific properties of the individual components and exhibit new fiber properties towards the outside. In order to prove that both proteins are actually involved in the formation of these novel fibrils, an analytical method is required that detects both sensitively and specifically on the nanometer scale. In recent years, tip-enhanced Raman spectroscopy (TERS) has proven to be very suitable in this field. The scanning of the sample at an accuracy within the nanometer range with the probe of a scanning force microscope (AFM) that is covered with a silver nanoparticle and simultaneous chemical characterization using enhanced Raman spectroscopy makes it possible to localize and classify both the secondary structure and individual amino acids on the fibrillar surface at an accuracy within the nanometer range. The crucial factor here is that the sample does not have to be provided with markers, which in turn can modify the fibril properties. In summary, molecular structural changes can be detected directly with a spatial resolution of only a few nanometers.
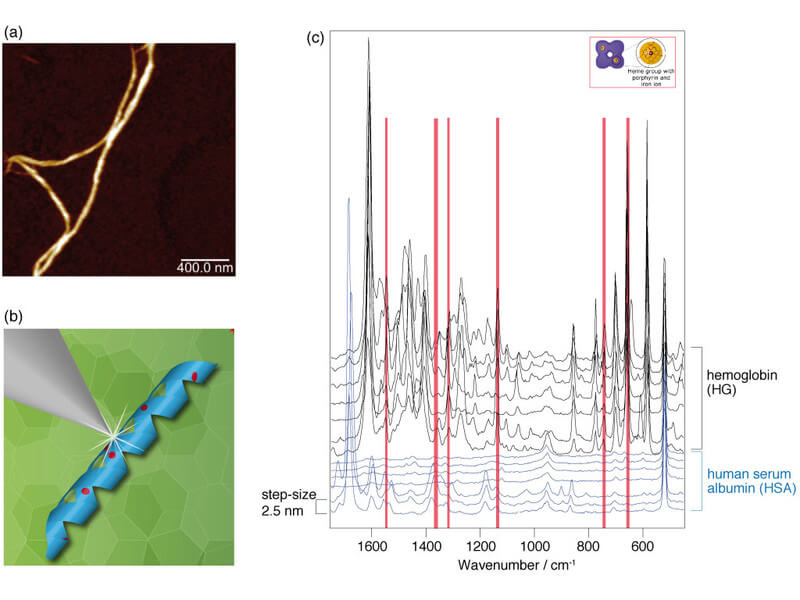
In TERS experiments on protein fibers synthesized from a mixture of hemoglobin (HG) and human serum albumin (HSA), the above method was used to demonstrate that both proteins are present in the fibers. The HG-HSA hybrid fibers were generated in ethanol solution at 65°C within seven days, passing through different agglomeration steps. The fibers for the TERS measurements were separated by application to a glass substrate. In the first step of the analysis procedure, the topography of the HG-HSA hybrid fibers was mapped (see Fig. 1a). Subsequently, TERS spectra were recorded at successive measuring points at an interval of 2.5 nm on the individual nanofibers (Figs. 1b and 1c). Due to the specificity and the high spatial resolution of the near-field method, hemoglobin and serum albumin could be unambiguously assigned using molecule-specific vibrational bands and localized on the fibers. Figure 1c shows an excerpt from a TERS measurement along an HG-HSA hybrid fiber in which the two proteins are easily distinguishable from each other. The first seven spectra can be assigned to HSA, while the following seven spectra have the characteristics of HG, which are determined by the bands of the heme group. Due to the resonant excitation of the HG molecule under the measuring conditions, the signal intensity is drastically increased compared to that of HSA. In all recorded data sets, the HSA fraction dominated, suggesting that the basic structure of the micrometer-long hybrid fibers consists of HSA, into which HG was partially incorporated. The TERS spectra also indicate that HG is irregularly distributed over the fibers. This assumption could be confirmed in a successful cooperation with the group of Prof. Jandt of the Otto Schott Institute for Materials Research of the FSU Jena by scanning electron microscopic measurements after immunogold marking of the samples.
The promising results of the study show that it is possible to produce nanofibers consisting of different proteins. This opens up the possibility of developing hybrid structures as new functional materials. One possible field of application for such structures is, for example, medical technology, in which biocompatibility plays a major role.
Funded by:
