- Home
- Research
- Spectroscopy and Imaging
- Research results
- Identification of exacerbation-relevant fungal spores in chronic respiratory diseases using UV Raman spectroscopy
Identification of exacerbation-relevant fungal spores in chronic respiratory diseases using UV Raman spectroscopy
28.03.2019
Exposure to particular species of fungal spores may lead to developing or worsening of the already present respiratory diseases. Thus, it is highly important to identify more dangerous species in the air. UV-Raman spectroscopy being able to provide spectral fingerprint of the microorganisms, has a potential accurately to assign the fungal spores to the correct genus, species or even strain level.
By O. Žukovskaja // K. Weber
Fungal spores are one of several environmental factors responsible for causing respiratory diseases like asthma, chronic obstructive pulmonary disease or aspergillosis and for triggering exacerbations for chronic forms of diseases. However, the disease and severity of exacerbation depends on fungal spores taxa and susceptibility of the person. Therefore, composition of the fungal spores in the air should be identified that susceptible individuals could perform preventions steps to avoid the exposure to more pathogenic fungal species. Currently the “gold standard” for fungal identification is based on the cultivation of sampled organisms. However, this laborious method is time consuming and requires highly trained personnel.
Raman spectroscopy can be an alternative tool for this task, since it provides information about molecular structure and chemical composition of samples. Different microorganisms vary, for example in the composition of the proteome or the cell wall, and therefore exhibit a slightly different Raman spectrum. Applying chemometric approaches, it is possible to capture even slight differences between the samples and to discriminate microorganisms at different taxonomic levels. However, for some pigmented species, it might be challenging due to high fluorescence background which overlaps with the Raman signal. Therefore, the application of UV-Raman spectroscopy with an excitation wavelength of 244 nm for the identification of fungal spores was investigated, since it allows to separate energetically the fluorescence and the Raman signal.
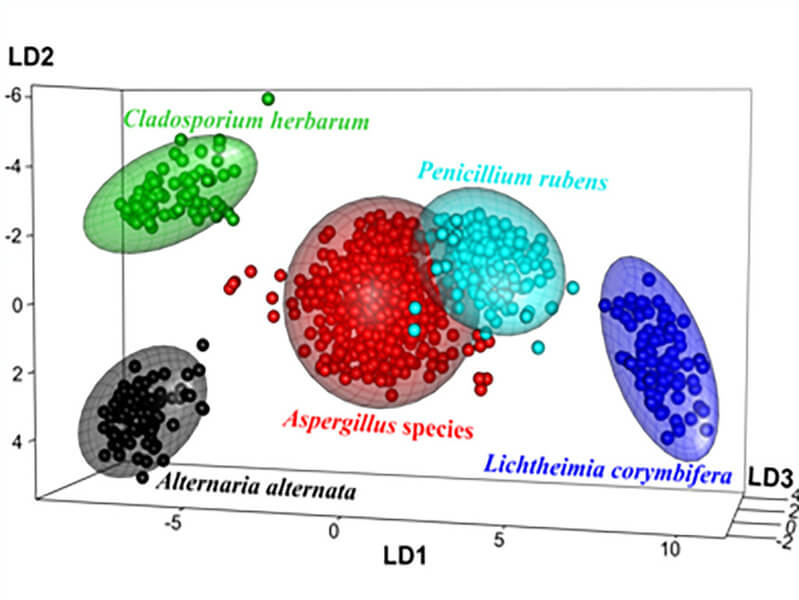
Three batches of eight different highly pigmented fungal species (Aspergillus fumigatus, Aspergillus niger, Aspergillus nidulans, Aspergillus calidoustus, Cladosporium herbarum, Alternaria alternata, Penicillium rubens, and Lichtheimia corymbifera), implicated to a varied extent in respiratory diseases worldwide, were measured using UV-Raman spectroscopy. Raman spectra of all species of fungal spores exhibit primary bands in the fingerprint region, which, in the case of UV-Raman spectroscopy, are typically associated with different nucleic acids and protein subunits. The spectral position of the bands was the same for all investigated species; however, intensities were varying, indicating different compositions of the spores. The collected spectra were used for the subsequent statistical analysis and classification models using Principal Component Analysis and Linear Discriminant Analysis were built and validated with leave-one-batch-out cross-validation.
Firstly, the classification model was created to distinguish spores on the genus level. The achieved results showed not only high accuracy of 97.5 % in classification, but also reflected the phylogenetic relationship between the different genera. A. alternataand C. herbariumbelonging to the same class of Dothideomycetes were nicely separated from Penicilliumand Aspergillusspores – assigned to the class of Eurotiomycetes, and from L. corymbiferaspores belonging to a lower group of fungi, the phylum Zygomycota (Figure 1). In the second step, the possibility to discriminate between different four Aspergillusspecies was investigated. The createdmodel assigned all spectra correctly resulting in an accuracy of 100 %. Finally, the model was trained to classify three different strains of A. fumigatus, since diversity in virulence among different A. fumigatusstrains is documented. The classification was correct for 89.4 % of spectra.
