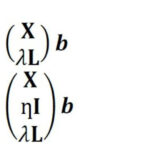- Home
- Research
- Photonic Data Science
- Research results
- Investigations on Raman Spectral Data Analysis
Investigations on Raman Spectral Data Analysis
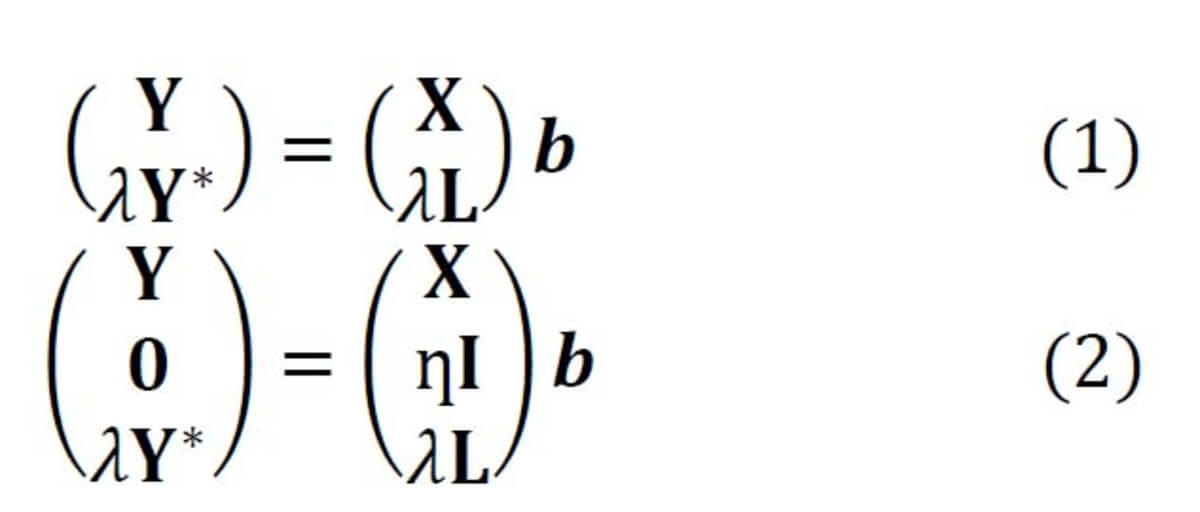
23.04.2018
If a Raman spectroscopy-based classification model is utilized to predict data measured on a different device or from a different biological replicate compared to the training data, the model’s performance is typically low. This problem can be tackled by transfer methods. We investigated Tikhonov regularization (TR)-based transfer methods and checked the model’s transfer performance.
By: Shuxia Guo // Ralf Heinke // Stephan Stöckel // Petra Rösch // Jürgen Popp // Thomas Bocklitz
Raman spectroscopy has been proven to be a versatile tool for biological studies including toxicology, microbiology, drug research, metabolic investigations, and forensic analysis [1]. These applications of Raman spectroscopy are only possible by using chemometrics for Raman spectral data analysis. Chemometrics improves the sensitivity of Raman-based biological detections, thanks to its capability of distinguishing subtle spectral variations caused by biological alterations. Therefore, we apply chemometrics in Raman-based biological studies and investigate possibilities of enhancing the outcome of the chemometric models. One of our investigated issues is the model transfer problem, which is required if the test data bears a significant difference to the training data due to variations in the different replicates or instruments. Model transfer approaches enable the trained model to successfully predict this new data without using a large amount of new training samples measured under the changed conditions. This is necessary especially if the new training samples are expensive or impossible to acquire, as in most biological studies. The training data is termed primary data, while the new data measured under different conditions is called secondary data.
In Raman spectroscopy, the most commonly applied model transfer method is standard calibration [2], in which the spectral variations between instruments are eliminated based on standardization measurements. However, the standard calibration does not have any effect on the variations in the replicates, which is one of the major causes of a failed prediction in biological applications. For this reason, we developed two approaches based on Tikhonov regularization (TR) [3], termed TR1 and TR2 [4]. The equations of these two approaches are given in Eq. (1-2), respectively. The method works by augmenting the training (primary) data (X,Y) with a few secondary training samples (L,Y*), which are called transfer spectra.
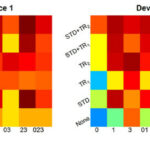
We based the investigation on the Raman spectra of three spore species (B. mycoides, B. subtilis, and B. thuringiensis) measured on four devices. A three-group classification model was constructed with a partial least square regression (PLSR). Each device was once used as secondary data and predicted by models trained with one or more of the other devices. In particular, for each case of secondary data, we randomly selected fifteen secondary spectra as optimization spectra, with five belonging to each group. The parameters λ and ƞ were optimized by genetic algorithms to maximize the prediction of these optimization spectra. The calculation was conducted on Raman spectra with or without standard calibration (STD). The prediction accuracy on the secondary data was visualized in Figure 1, which was significantly improved after model transfer (i.e., compared to the prediction without model transfer). Both methods, namely TR1 and TR2, were able to significantly improve the prediction, even without carrying out a standard calibration. Nonetheless, TR2 is superior to TR1.
Funded by: China scholarship council, BMBF, EU